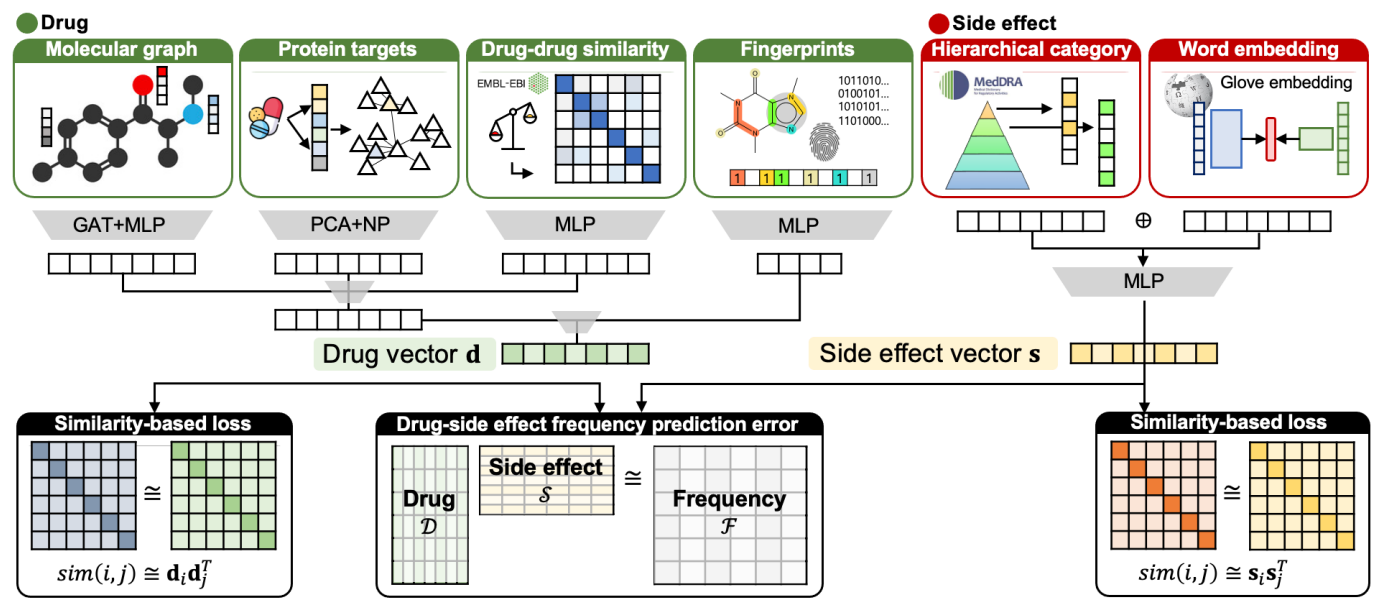
Deep Learning models for interpreting biological data with prior knowledge
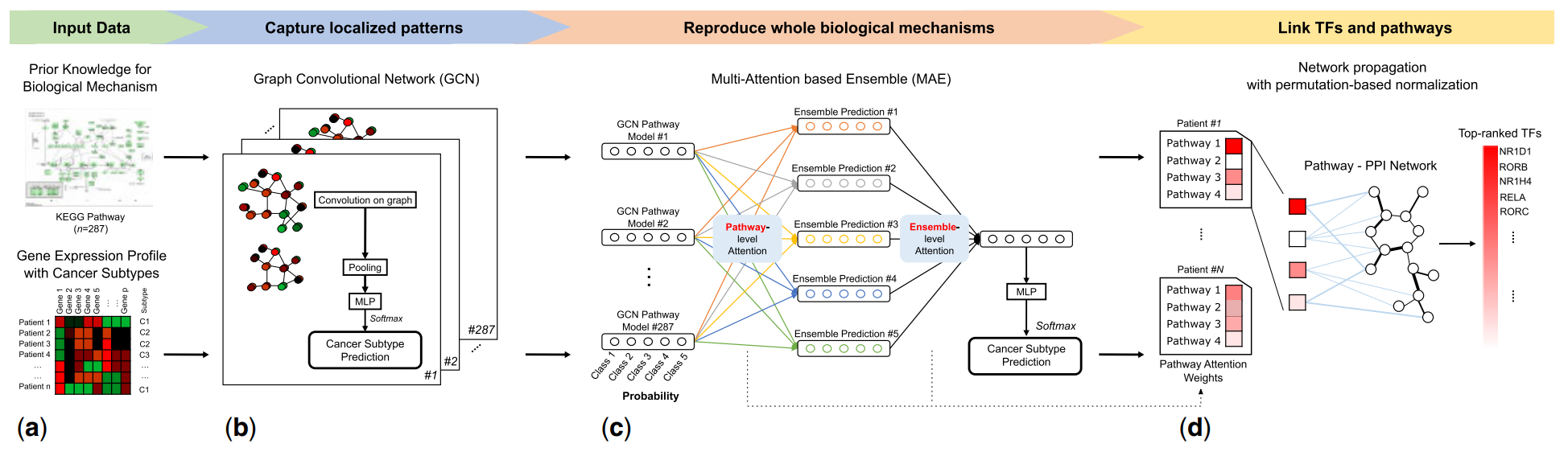
Cancer subtype classification and modeling by pathway attention and propagation
Designing a pathway-based explainable deep learning model by graph convolutional network and attention mechanism
Predicting cancer subtypes using gene expression data and pathway information
Paper accepted by Bioinformatics journal
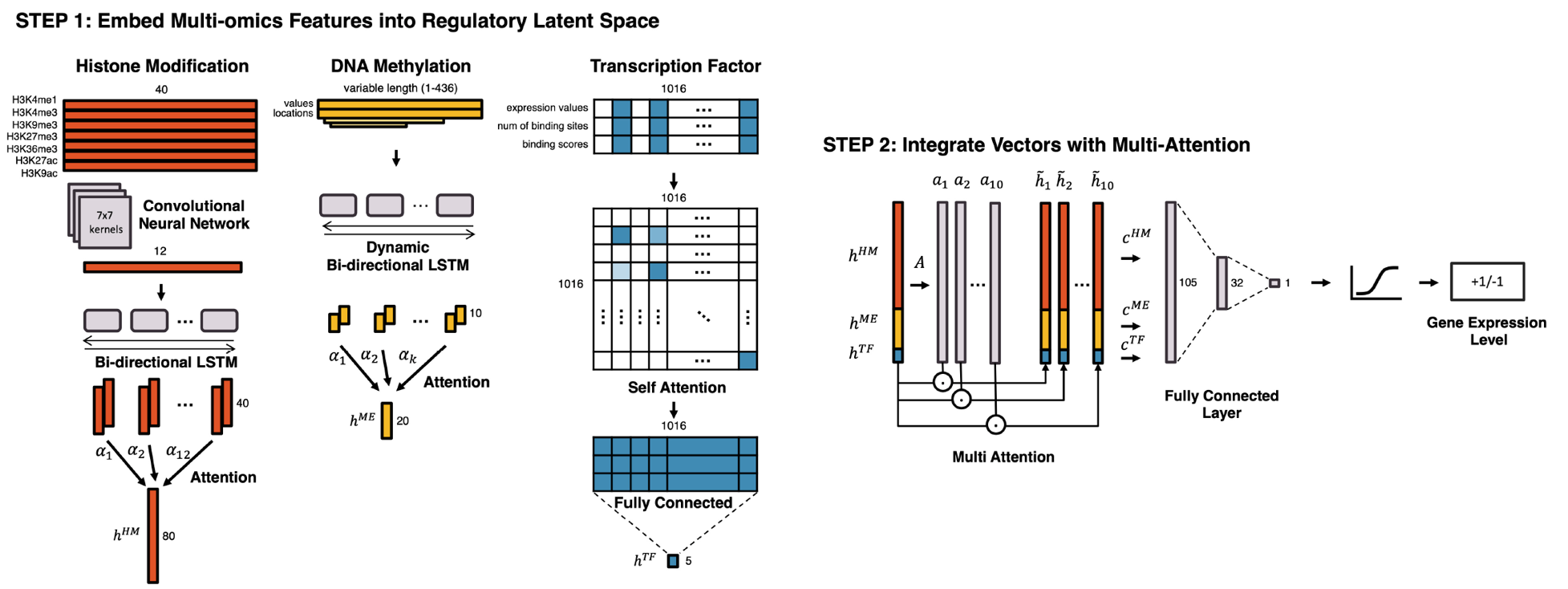
Learning Cell-Type-Specific Gene Regulation Mechanisms by Multi-Attention Based Deep Learning with Regulatory Latent Space
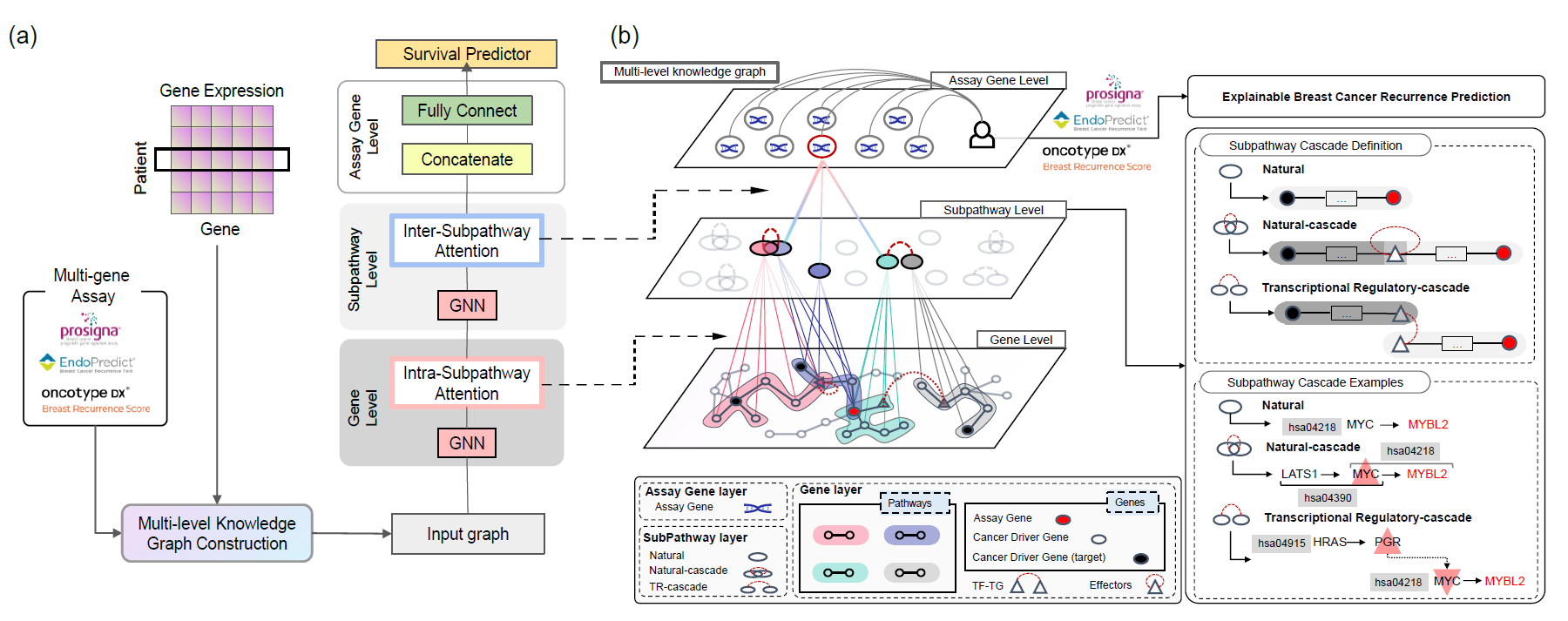
Multi-layered Knowledge Graph Neural Network Reveals Pathway-level Agreement of Three Breast Cancer Multi-gene Assays
Designing an explainable deep learning model by integrating intra- and inter- pathway level attention
Identification of shared regulatory mechanisms of three breast cancer multi-gene assays
Under review in SCIE journal
AI with graph structured data in biomedical domains
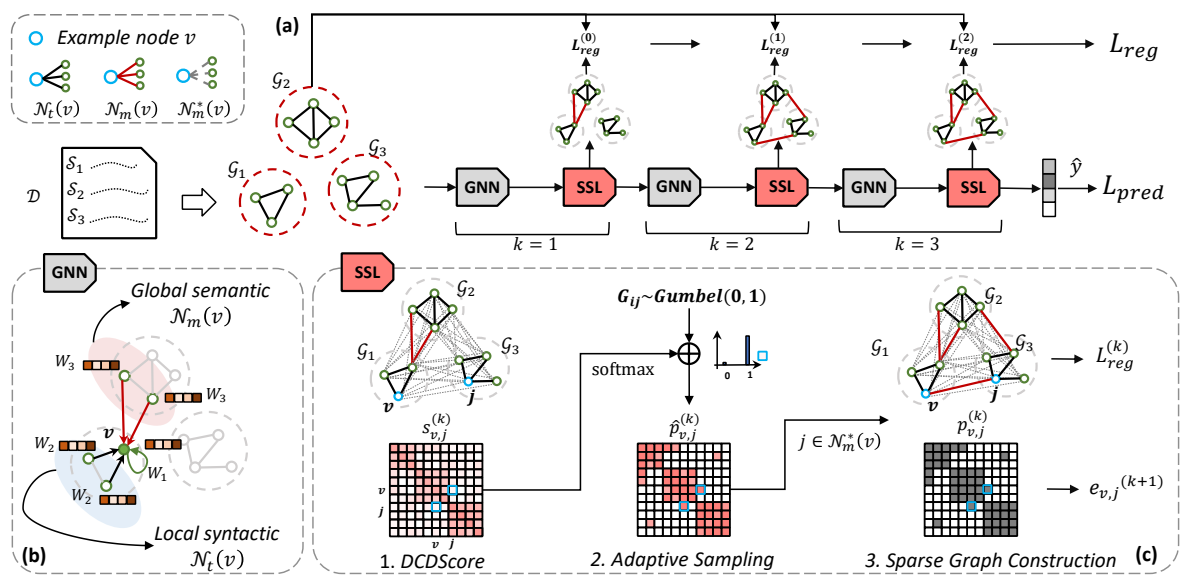
Sparse Structure Learning via Graph Neural Networks for Inductive Document Classification
A novel GNN-based sparse structure learning model for inductive document classification
Employing structure learning to sparsely select edges between words by considering dynamic contextual dependencies
Paper accepted by AAAI 2022
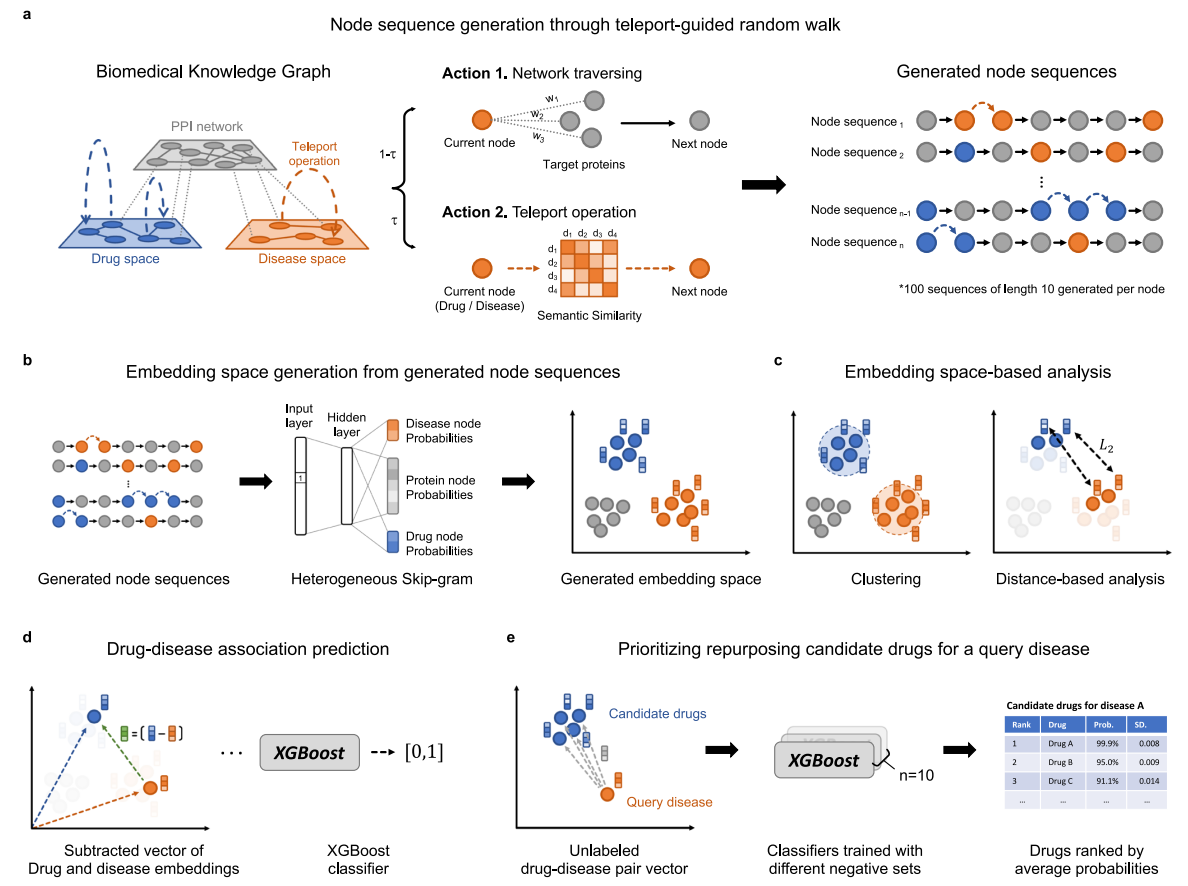
Biomedical knowledge graph learning for drug repurposing by extending guilt-by-association to multiple layers
A semantic multi-layer guilt-by-association approach that leverages the principle of guilt-by-association - “similar genes share similar functions” at the drug-gene-disease level
Designing a semantic information-guided random walk to generate embeddings of drugs and disease in a unified embedding space
Paper accepted by Nature Communications journal
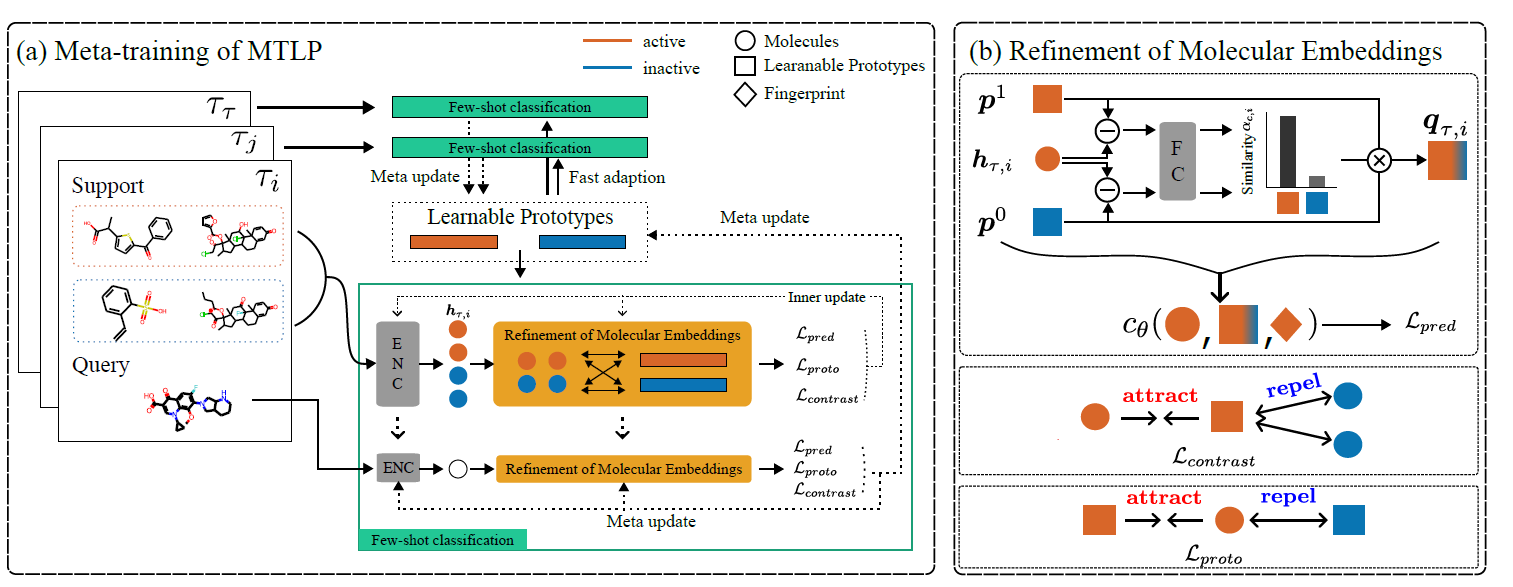
Leveraging shared knowledge across multiple molecular properties in few-shot learning
Incorporating a stochastic attention mechanisms to reflect information from multiple assays
Under review in International Conference
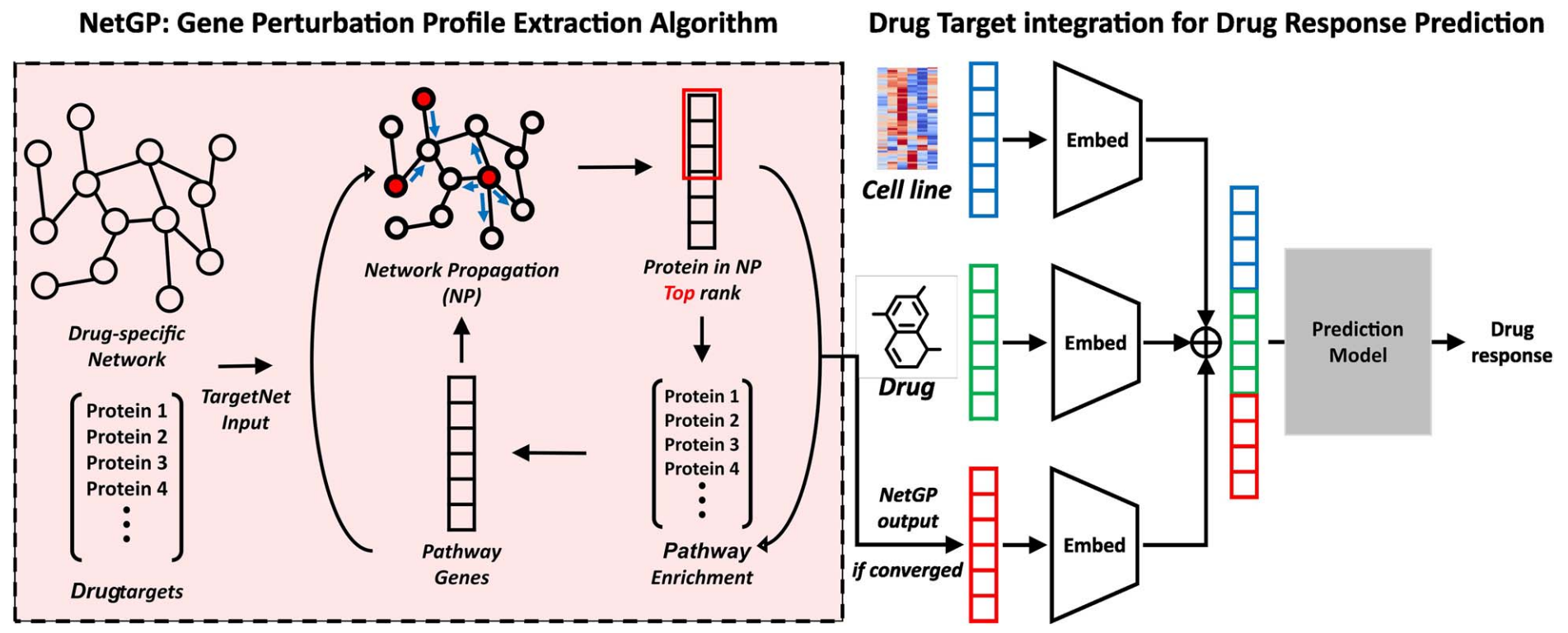
Improved drug response prediction by drug target data integration via network-based profiling